Products & Applications
Sequins WGS Clinical Control Set
Maximize insights from every sample with Sequins™ internal standards.


Most Comprehensive WGS Clinical Control Set
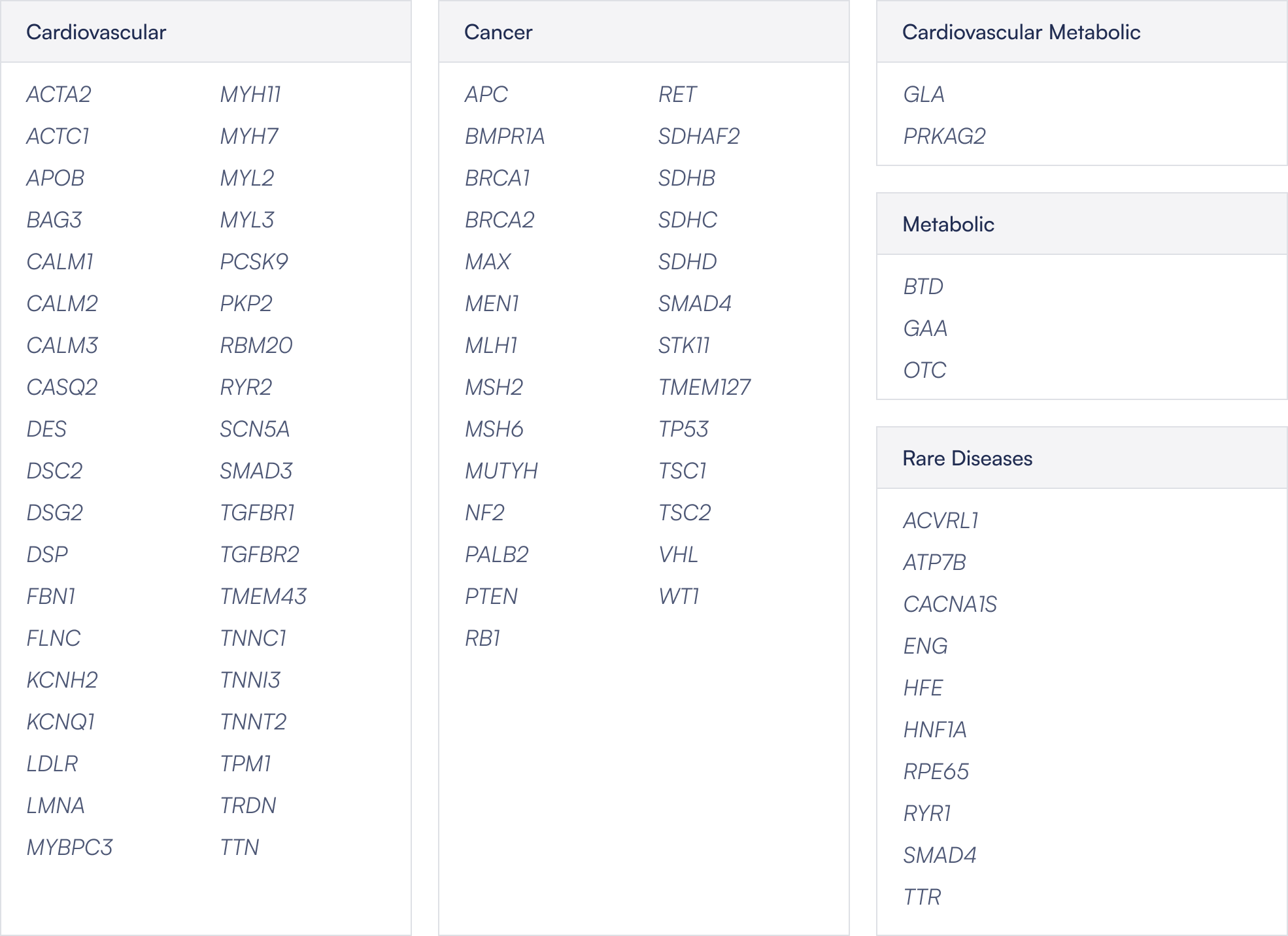
Broad Coverage: Sequins representing 80 of the 84 clinically actionable ACMG Secondary Findings genes across cancer, cardiovascular, metabolic, and rare diseases.
Flexible and Compatible: Available as a standalone kit or in combination with the WGS Core Control Set for an integrated control system.
Comprehensive Variant Representation: Covers pathogenic /likely pathogenic, VUS and benign/likely benign variants in ACMG-listed secondary findings genes, enabling robust benchmarking of variant detection and reporting pipelines.
Gene list

WGS Clinical Control Set v1.0 Performance
To assess performance of Sequins relative to native human genomic DNA (hgDNA), we compared read coverage across matched Sequins target regions. Sequins from the WGS Clinical Control Set v1.0 demonstrated strong concordance with hgDNA, and this relationship was preserved when combined with the WGS Core Control Set. These results confirm that Sequins recapitulate native sequence behaviour and can be integrated across control modules without compromising coverage.
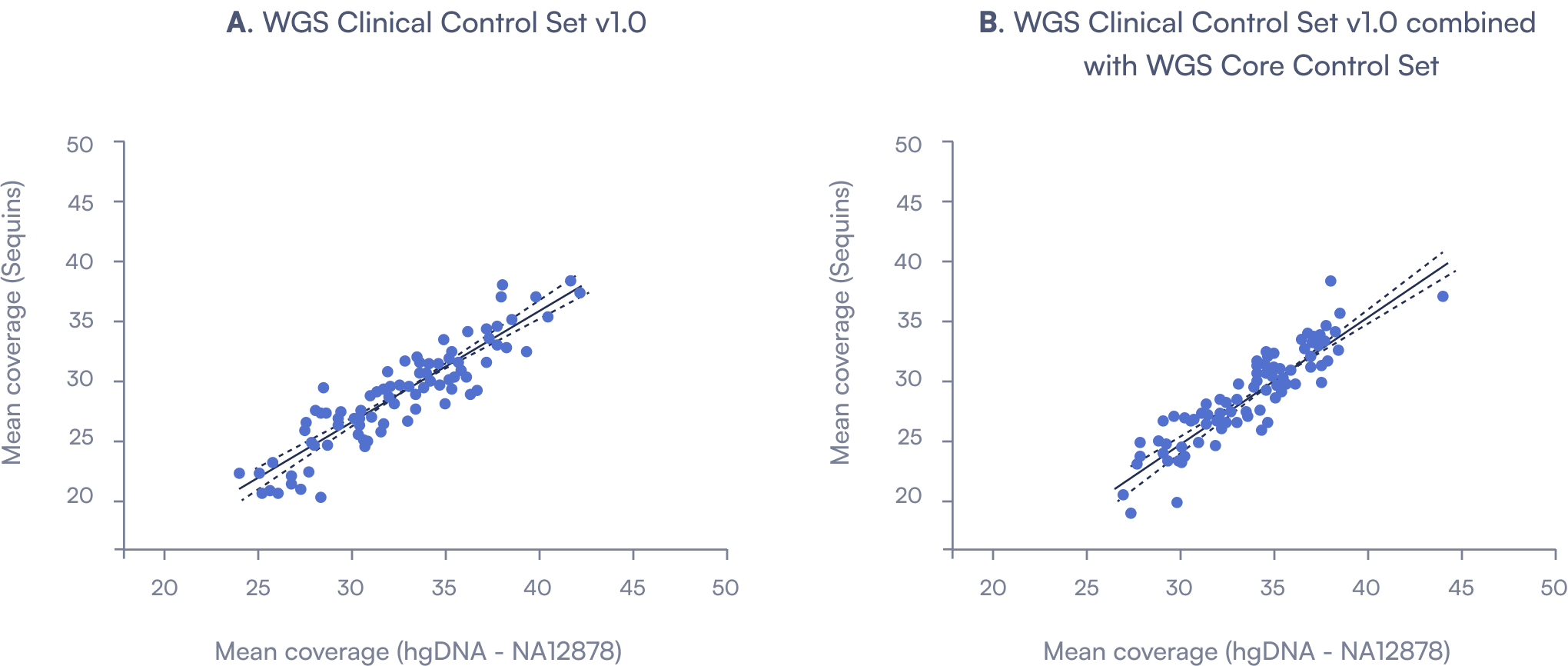
Mean coverage of Sequins and hgDNA at the same location. (A) Reads mapped to the Sequins WGS Clinical Control Set v1.0 strongly correlated with the same regions of hgDNA (slope 0.89, R² = 0.83). (B) This correlation was maintained when combined with the WGS Core Control Set (slope 1.00, R² = 0.82), confirming compatibility without loss of performance.

Variant calling across the full genome of NA12878 showed precision and sensitivity above 99% in all conditions. Importantly, combining the WGS Clinical Control Set v1.0 with the WGS Core Control Set did not alter the accuracy of hgDNA variant calls. The observed differences were within the expected background noise of large-scale variant calling pipelines.
| Sample | TP | FP | FN | Precision | Sensitivity | F-measure |
|---|---|---|---|---|---|---|
| NA12878 + WGS Clinical Control Set v1.0 | 3,682,018 | 28,890 | 18,083 | 0.9922 | 0.9951 | 0.9937 |
| NA12878 + WGS Clinical Control Set v1.0 + WGS Core Control Set | 3,682,079 | 28,849 | 18,009 | 0.9922 | 0.9951 | 0.9937 |
TP – true positive, FP – false positive, FN – false negative
Within the Sequins target regions, variant calling performance was near-perfect. Precision reached 100%, sensitivity exceeded 99.7%, and the only false negative was an expected structural variant not detectable by the germline variant caller used. Results were identical when combined with the WGS Core Control Set.
| Sample | TP | FP | FN* | Precision | Sensitivity | F-measure |
|---|---|---|---|---|---|---|
| NA12878 + WGS Clinical Control Set v1.0 | 385 | 0 | 1 | 1.0000 | 0.9974 | 0.9987 |
| NA12878 + WGS Clinical Control Set v1.0 + WGS Core Control Set | 385 | 0 | 1 | 1.0000 | 0.9974 | 0.9987 |
TP – true positive, FP – false positive, FN – false negative
*The single false negative was expected due to limitation of the variant caller for structural variant




