Products & Applications
Sequins Metagenomics Core Control Set
Industry leading quantification standards for microbial populations.


Most Comprehensive Metagenomics Control Set
The Sequins™ Metagenomics Core Control Set is a comprehensive, easy-to-use, pre-configured control set with built-in redundancy that ensures integrity of results. It contains synthetic sequences that collectively represent 52 microbial species at varying abundances to emulate the complexity found in a natural microbial community. Features include: quantitative ladders; wide representation of domains including bacteria, archaea and eukaryotes; GC% content range (27% to 71%); single tube blend; demonstrated on various samples using Illumina and Oxford Nanopore technologies.
| Genus | No. Species | Genus | No. Species | Genus | No. Species |
| Aciduliprofundum | 1 | Fusobacterium | 1 | Persephonella | 1 |
| Acinetobacter | 2 | Helicobacter | 1 | Porphyromonas | 1 |
| Anabaena | 1 | Klebsiella | 1 | Roseobacter | 1 |
| Bacillus | 2 | Lactobacillus | 1 | Salmonella | 1 |
| Buchnera | 1 | Legionella | 1 | Shigella | 1 |
| Caldicellulosiruptor | 1 | Leuconostoc | 1 | Staphylococcus | 1 |
| Candida | 1 | Listeria | 2 | Streptococcus | 1 |
| Candidatus Caldiarchaeum | 1 | Magnetococcus | 1 | Synechocystis | 1 |
| Candidatus Carsonella | 1 | Metallosphaera | 1 | Thermotogan | 1 |
| Candidatus Korarchaeum | 1 | Methanococcus | 1 | Thermus | 1 |
| Chlamydia | 1 | Methanopyrus | 1 | Toxoplasma | 1 |
| Chlorobium | 1 | Mycobacterium | 1 | Treponema | 1 |
| Corynebacterium | 1 | Neisseria | 2 | Vibrio | 1 |
| Desulfitobacterium | 1 | Nitrosopumilus | 1 | Vulcanisaeta | 1 |
| Desulfovibrio | 1 | Nostoc | 1 | Wolinella | 1 |
| Ehrlichia | 1 | Oenococcus | 1 | Yersinia | 1 |


Best In-Class Quantification & LOD Determination
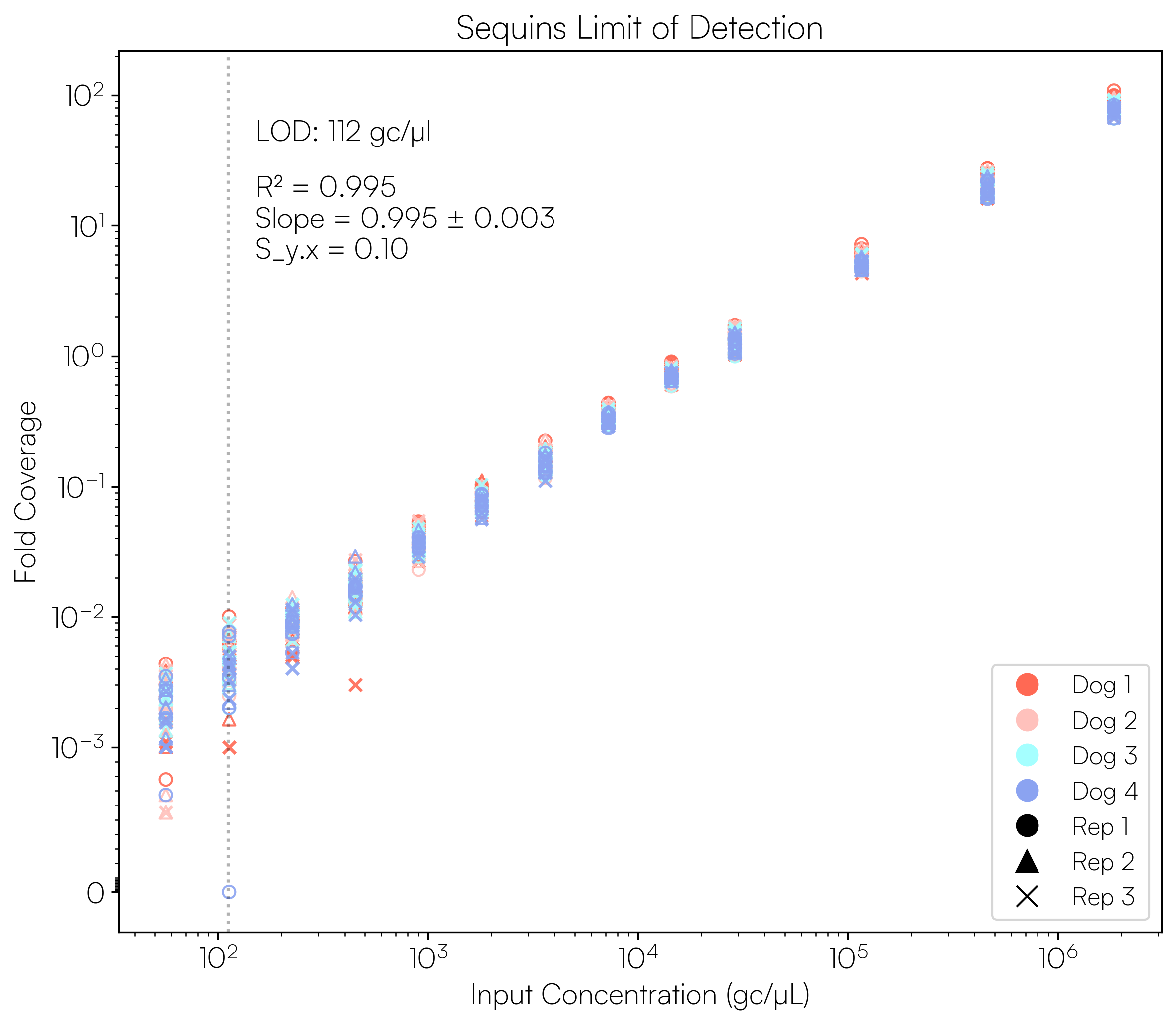
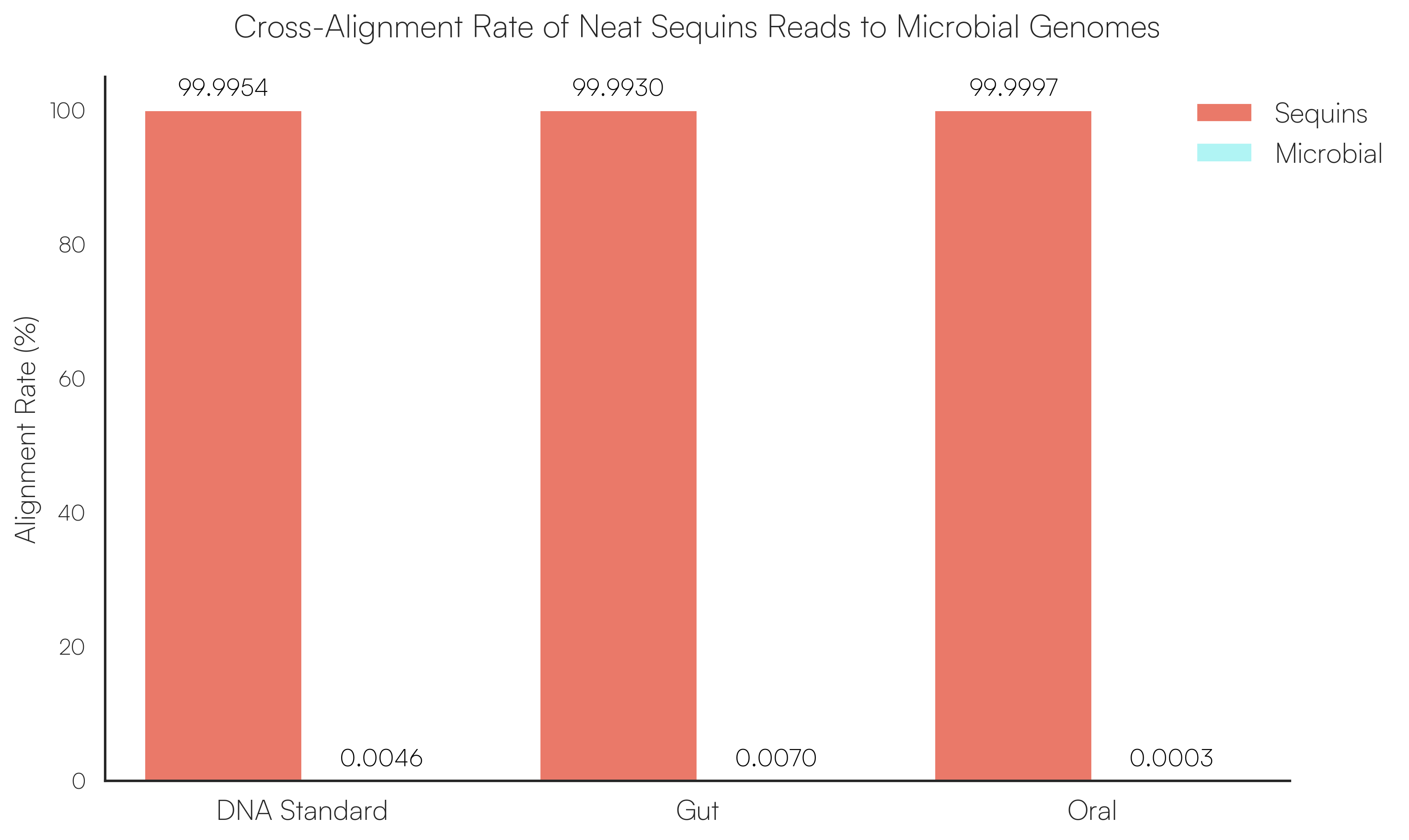
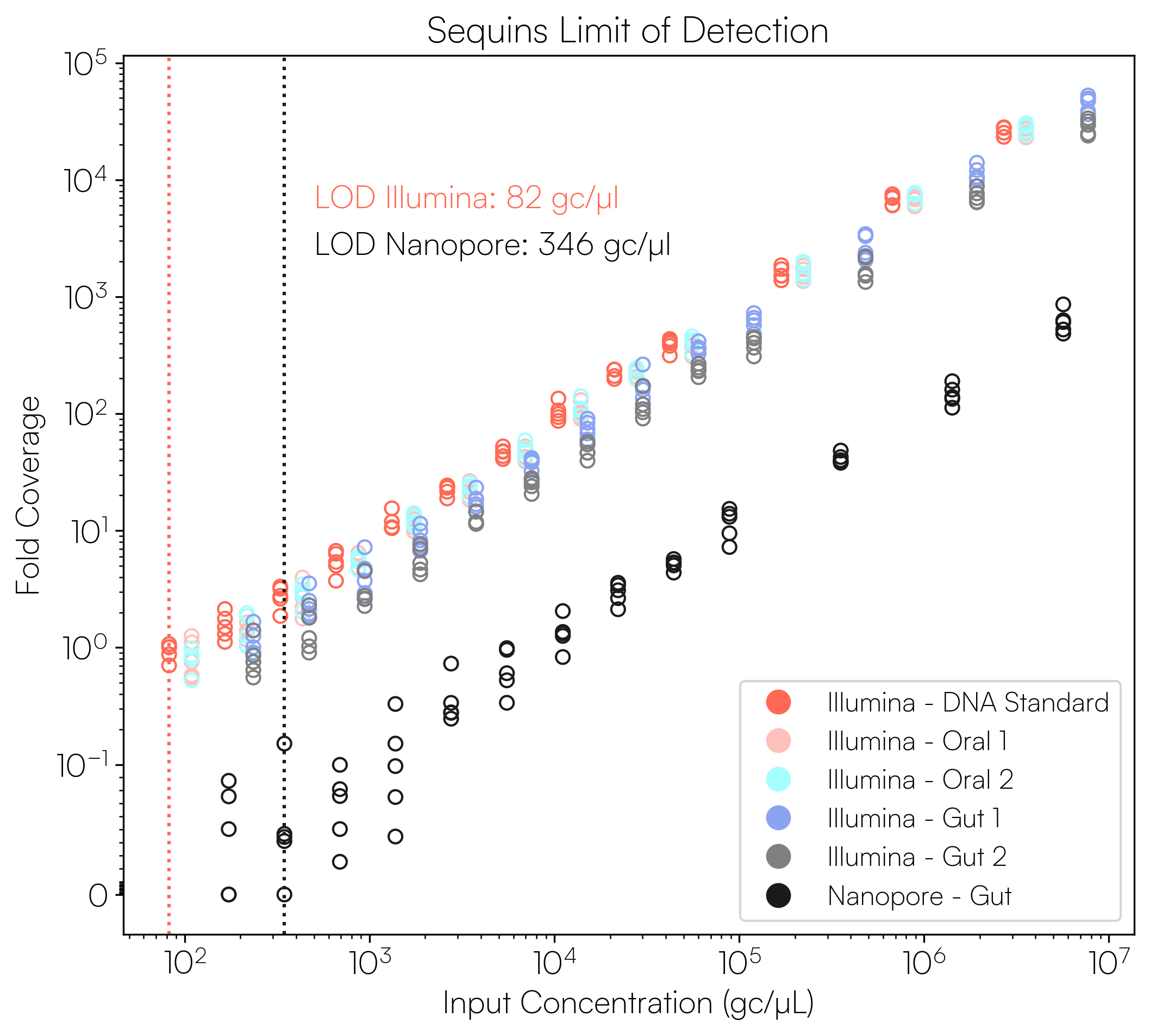
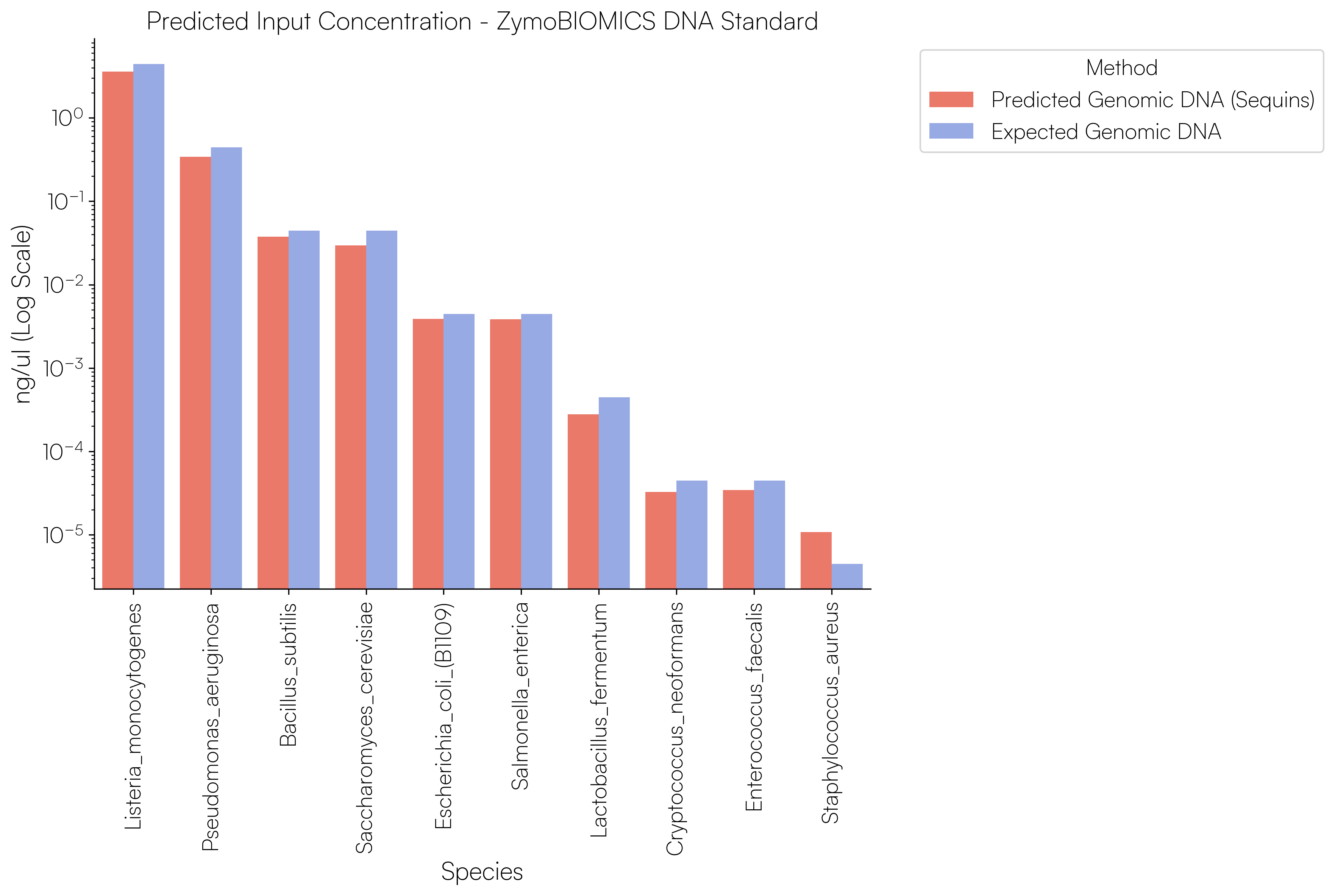
Sequins Metagenomics Core Control Set performance. (A) Metagenomics Core Control Set replicates in dog faecal matrix background demonstrating an accurate and reproducible ladder in complex samples. (B) Cross-alignment rate of Sequins reads to microbial genomes, demonstrating that Sequins does not interfere with samples – less than 0.01% of reads cross-align to mock community genomes. (C) Sequins ladder on various backgrounds generated using Illumina and Oxford Nanopore Technologies (ONT) sequencing. This demonstrates that Sequins is compatible with short- and long-read sequencing technologies and can be used to determine assay limit of detection (LOD). (D) Predicted input concentration estimates (predicted using Sequins v. expected based on ZymoBIOMICS DNA Standard) demonstrating that the Sequins molecular ladder enables reliable microbial abundance quantification. Internal data.


Best In-Class Normalization
Sequins provide superior normalization performance. Relative Log Expression (RLE) normalization using (A) raw reads counts, (B) upper quartile and (C) Sequins. Faecal samples collected from Dogs 1 to 4, each with three corresponding replicates (DP1-DP12). Sequins-based normalization showed the most pronounced improvements. This included a tighter distribution centred around zero and reduced inter-sample variation. All results are based on phylum-level abundance estimates from Kraken 2/Bracken.
![Detailed plot displaying measurement values across samples: the x-axis shows [X variable], the y-axis shows [Y variable], and cell/marker color indicates value intensity; large axes labels and a legend are included to aid interpretation.](https://sequins.bio/wp-content/uploads/2025/04/image002.png)