Products & Applications
Sequins ctDNA Evaluation Set
A new era of ctDNA accuracy: powerful calibration built into every sample.


Circulating tumor DNA (ctDNA) sequencing has fast become a powerful method for early detection, screening and recurrence monitoring of cancer. Liquid biopsies for ctDNA can quickly detect cancer-associated actionable variants, reveal information about cancer type and stage and response to therapies.
ctDNA is typically present at very low variant allele frequencies (VAFs) making detection accuracy dependent on high sequencing coverage, high fragment depth, and assay calibration.
Sequins internal standards enable precise estimation of VAFs, determination of the limit of detection (LoD), assay normalization, and calibration of variant calls against internal benchmarks.

Customizable Control Backbone with Molecular Ladder
The Sequins ctDNA Evaluation Set comprises a molecular ladder developed for the application of assessing variant allele frequency and sensitivity in the context of general cell-free cancer testing. The set targets twenty-two cancer genes with seventeen challenging INDELs and five SNPs at precise variant allele frequencies from 0.1 to 10% and is applicable to a range of cancers.
| Gene | ClinVar UID | Variant | VAF % |
|---|---|---|---|
| BRCA1 | ClinVar_1076672 | c.2219_2220insTAAT (p.Ser741fs) | 0.1 |
| WT1 | ClinVar_449416 | c.1120C>T (p.Arg374Ter) | 0.1 |
| SMAD4 | ClinVar_24867 | c.1587dup (p.His530fs) | 0.1 |
| APC | ClinVar_1072211 | c.476_488del (p.Tyr158_Tyr159insTer) | 0.1 |
| RB1 | ClinVar_1073997 | c.46_61GCC[2]GCGGAACCCCAGGCACCGCCGCCGCCGCCGCCGCGGAACCCC[1] (p.Pro21fs) | 0.1 |
| BRCA2 | ClinVar_9342 | c.658_659del (p.Val220fs) | 0.5 |
| PMS2 | ClinVar_1328224 | c.741_742insGTGTGTGAAG (p.Ser248fs) | 0.5 |
| TSC1 | ClinVar_1013340 | c.903dup (p.Asn302fs) | 0.5 |
| VHL | ClinVar_2218 | c.499C>T (p.Arg167Trp) | 0.5 |
| MEN1 | ClinVar_1070954 | c.1375_1391dup (p.Ala467fs) | 0.5 |
| MAX | ClinVar_404110 | c.211_221del (p.Ile71fs) | 0.5 |
| MLH1 | ClinVar_89935 | c.18_34del (p.Val7fs) | 1.0 |
| PTEN | ClinVar_545882 | c.865_866insTTCT (p.Lys289fs) | 1.0 |
| SDHAF2 | ClinVar_532513 | c.177dup (p.Asp60Ter) | 1.0 |
| TSC2 | ClinVar_237966 | c.148A>G (p.Met50Val) | 1.0 |
| MSH6 | ClinVar_1070916 | c.3964_3980dup (p.Asn1327delinsLysAsnLeuArgArgTer) | 5.0 |
| BMPR1A | ClinVar_529927 | c.366_384del (p.Glu123fs) | 5.0 |
| TMEM127 | ClinVar_126966 | c.265_268del (p.Thr89fs) | 5.0 |
| MSH2 | ClinVar_90711 | c.1638_1639dup (p.Asn547fs) | 10.0 |
| RET | ClinVar_230926 | c.1998G>C (p.Lys666Asn) | 10.0 |
| STK11 | ClinGen_CA402950689 | g.1221314G>A (g.1221314G>A) | 10.0 |
| SDHB | ClinVar_428926 | c.17_42dup (p.Ala15delinsProSerProTer) | 10.0 |
The Sequins ctDNA Evaluation Set can be used as a backbone for further customization with additional gene targets through our Sequins On-Demand program.


Sequins Performance
Molecular ladder maintains linearity across all spike-in amounts.
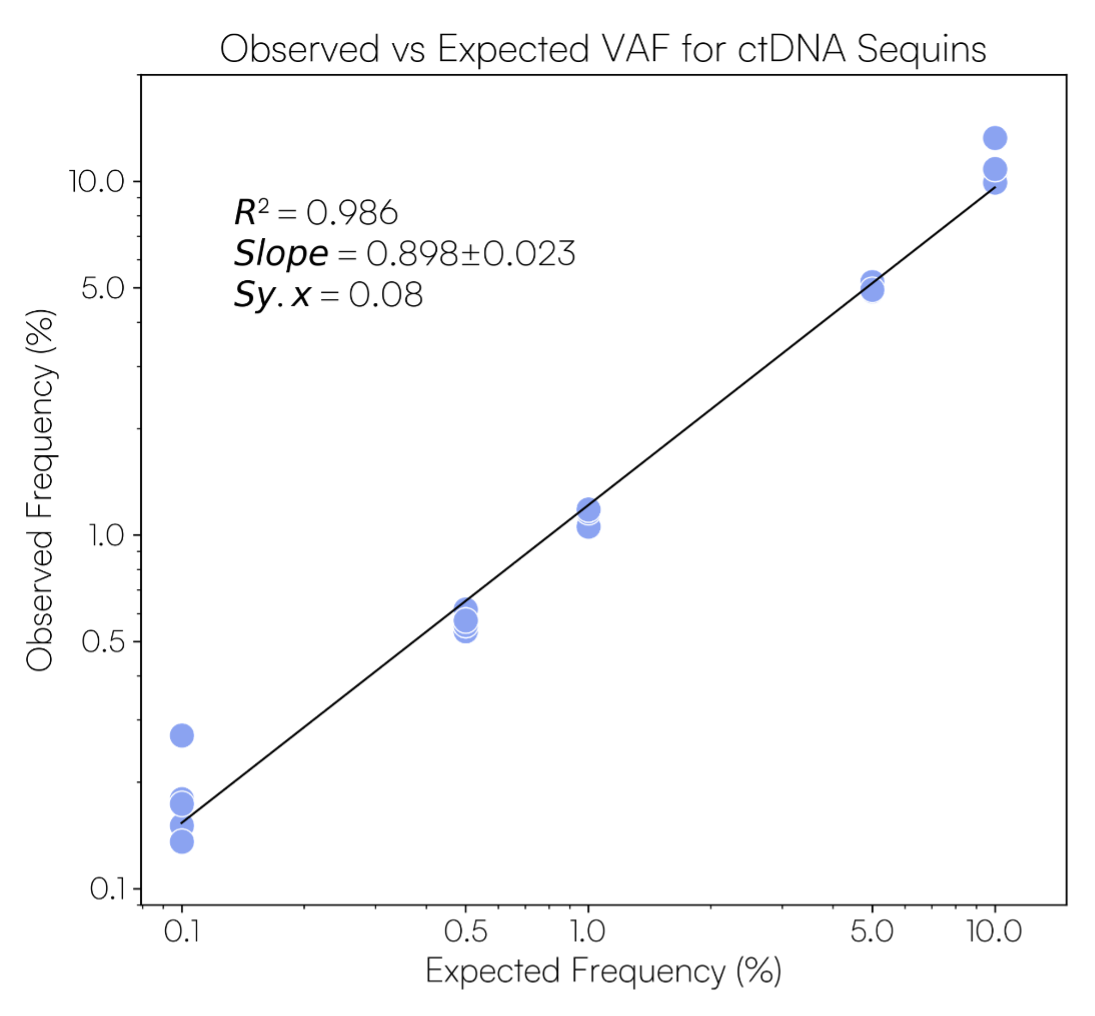
Observed versus expected variant allele frequencies (VAFs) for synthetic Sequins variants. Twenty-two cancer gene variants representing varying allele frequencies (0.1%, 0.5%, 1%, 5%, 10%) were sequenced neat on Illumina NextSeq platform using IDT xGen cfDNA & FFPE Library prep kit v2 MC.


Compatibility with Existing Panels
The Sequins Control Probe Set showed no interference with the IDT Human ID Hyb Panel. Coverage uniformity across targets remained consistent whether captured alone or co-captured with the Sequins Control Probe Set and varying Sequins spike-in levels.
Apply Now

Designed to be run in combination with targeted gene panels, users can source a vendor to design and synthesize capture probes for the Sequins ctDNA Evaluation Set.
Provides a comprehensive backbone and can be further customized with additional gene targets through our On-Demand program.
We are releasing the Sequins ctDNA Evaluation Set through our Technology Access Program.




